The Evolution of Antimicrobial Resistance
The term "anti-microbial" is a general term that encompasses drugs, chemicals, or other substances that either kill or slow the growth microbes. These include:
- Anti-bacterial drugs (antibiotics), which kill bacteria
- Anti-viral agents, which kill viruses
- Anti-fungal agents, which kill fungi, and
- Anti-parisitic drugs, which kill protozoa
Scottish scientist Alexander Fleming is widely credited with the discovery of the antibiotic properties of penicillin in 1928, although an earlier report had noted the ability of penicillin mold to kill bacteria as early as 1897. There are scattered reports of penicillin mold being used to treat gonococcal infections of the eye of newborns as early as 1930, but it wasn't until the 1940s that penicillin began to be used for clinical infections. It was soon recognized to be a truly remarkable drug. Small doses cured infections caused by Staphylococcus, Streptococcus, Neisseria, syphilis, and many other bacteria. Penicillin use became increasingly widespread and indiscriminate, and resistant strains of bacteria emerged. Increasingly large doses of pencillin were require to cure infections, and some strains were entirely resistant. Other antibiotics were developed, and synthetic penicillin-like drugs were introduced, but strains of bacteria resistant to these newer antibiotics also emerged. At first, these problems were dismissed, but by the 1980s, it had become clear that antibiotic resistance was an important and growing problem. In the 1950s studies were published showing that animals given low doses of antibiotics gained weight more rapidly, and the practice of including antibiotics in grain to promote the growth of cattle, poultry, and swine became widepspread, further compounding the problem of bacterial resistance to antibiotics.
Antibiotics kill or inhibit the growth of susceptible bacteria. Sometimes one of the bacteria survives because it has the ability to neutralize or evade the effect of the antibiotic; that one bacteria can then multiply and replace all the bacteria that were killed off. Exposure to antibiotics therefore provides selective pressure, which makes the surviving bacteria more likely to be resistant. In addition, bacteria that were at one time susceptible to an antibiotic can acquire resistance through mutation of their genetic material or by acquiring pieces of DNA that code for the resistance properties from other bacteria. The DNA that codes for resistance can be grouped in a single easily transferable package. This means that bacteria can become resistant to many antimicrobial agents because of the transfer of one piece of DNA.
The iFrame below is a CDC web page that provides brief descriptions of the many infectious agents that have developed drug resistance.
For more information on antimicrobial resistance due to antibiotic treatment of livestock see the Antimicrobial Resistance Learning Site.
Penicillin-Resistant Bacteria
Penicillin's ability to kill bacteria was due to it's ability to inhibit a bacterial enzyme that was essential for synthesis of the bacterial cell wall. Penicillin's ability to do this depended on a key structure called a "β-lactam ring". Resistance to penicillin initially occurred as a result of a mutation in a bacterium that created an enzyme (penicillinase) which was capable of breaking down the β-lactam ring. Bacteria that possessed this resistance first evolved in hospitals, but they rapidly spread to the wider community at large. The gene encoding for penicillinase resided not on the bacterial chromosome, but on an extra circlular ring of DNA referred to as a "plasmid." This extra piece of bacerial DNA can be replicated and transferred from a resistant bacterium to one that was previously susceptible by a process referred to as bacterial conjugation.
Methicillin-Resistant Staphylococcus aureus (MRSA)
Methicillin, a chemically modified version of penicillin, was introduced in 1959 to treat infections caused by bacteria resistant to penicillin, but it was effective against a narrower spectrum of bacteria. In addition, strains of Staphylococci resistant to methicillin were reported as early as 1961; these strains had acquired a gene (mecA) wihich inacitivates methicillin by encoding for a protein that binds to it. The mecA gene is carried on an extra "mobile genetic element", the staphylococcal cassette chromosome (SCCmec). Initially, MRSA strains were encountered only in hospitals, but in the late 1990s MRSA was found in the community at large and quickly spread worldwide.
Students in PH709 created a 7-minute public service announcement that explains how MRSA evolved in hospitals and eventually escaped to the community to cause cases of so-called "community-acquired MRSA."
You can learn more about the issues surrounding MRSA and measures to control is by exploring iFrame below which links to the CDC web page on MRSA.
Bacterial Acquistion of Antibiotic Resistant Genes
As we noted above, novel genes arise from random mutations, and occasionally such a mutation may confer a bacterium with resistance to an antiobiotic. Once a bacterium has acquired resistance to a particular antibiotic, it passes the resistant allele to subsequent daughter cells that result from binary fission. In addition, bacteria that have acquire a trait such as antibiotic resistance can transfer this allele to other bacteria throught any one of three mechanisms:
- Transformation
- Conjugation
- Transduction
Transformation
When bacterial cells die, they frequently lyse (burst) releasing their intracellular contents, including fragments of DNA, to the environment. These fragments can be taken up and incorporated into the chromosome of a living bacterium to provide the recipient with new characteristics. This process is called bacterial transformation, and if the incorporated DNA contains genes that encode for resistance to an antibiotic, a previously susceptible bacterium can be "transformed" to now be resistant. The video below (23 sec.) provides a quick overview of transformation.
Conjugation
Many bacteria have plasmids, which are small circular pieces of DNA separate from the primary bacterial chromosome. These plasmids can carry genes that provide resistance to antibiotics, and bacteria that contain plasmids are able to conjugate with other bacteria and pass a replicate to recipient bacteria. The electron micrograph below shows two bacteria that are joined by a temporary hollow tube-like connection called a pilus.

Source: http://evolution.berkeley.edu/evolibrary/article/side_0_0/turboevolution_01
The video below (2 min 15 sec) provides an excellent overview of conjugation.
In 1968 a Shigella epidemic killed 12,500 people in Guatemala. The Shigella bacteria that caused the outbreak had a plasmid carrying resistances to four antibiotics.
Transduction
Genetic information can also be carried from one bacterium to another by a virus. Bacteriophages (or simply "phages") are small viruses that infect bacteria and use their cellular components to make bacteriophage replicates. During the infection and replication, it is possible for bacterial genes to get incorporated into the viral genome. One of the viral replicates carrying the bacterial allele may then subsequently infect another bacterium and pass the new allele on.
The illustration below begins with a donor bacterium that has a gene (a+) which encodes for resistance to a particular antibiotic. In the 2nd and 3rd scenes the viral proteins and genetic material are replicated and then self-assemble into new viral particles. In the process, however, the bacterial gene for a+ gets incorporated into one of the virus particles. The viral particles are then released from the donor bacterium, and the virus with the a+ gene infects a recipient bacterium, after which the a+ gene can become incoporated tinto the genome of the recipient, which has now acquired this gene for antibiotic resistance.
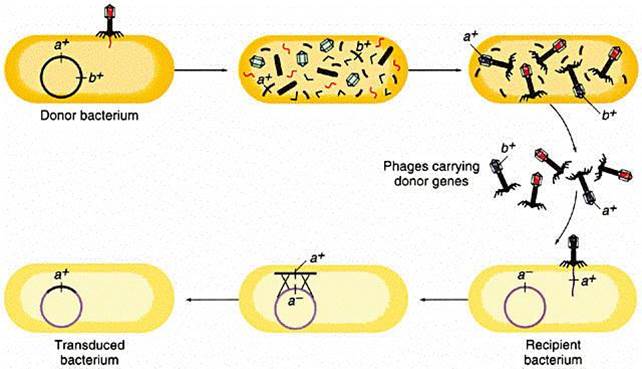
Source: http://www.cbs.dtu.dk/courses/genomics_course/roanoke/genetics980309.html
Slowing the Phenomenon of Antibiotic Resistance
The development and spread of bacterial resistance to antiobiotics is inevitable, but it could be greatly curtailed through relatively simple measures. These include:
- Preventing infection (general infection control)
- Hand washing among health care workers, food handlers, and the general public
- Modern sanitation: effective systems for dealing with sewage & providing clean water
- Proper food preparation practices
- Rapid identification & isolation of new cases of infection, e.g., new cases of TB; this is particularly important with drug-resistant cases
- Continued development of new antibiotics
- Decreased agricultural use of antibiotics to enhance growth
- Physician education to reduce inappropriate prescriptions & inappropriate use of broad-spectrum antibiotics
- Educating physicians and patients about the importance taking the appropriate dose of an antibiotic for the full period of treatment that is indicated
- Consumer education regarding the importance of bacterial resistance and the uselessness of taking antibiotics for viral infections such as the common cold (See the CDC's Education Campaigns to combat microbial resistance.)
See also the CDC's Public Health Action Plan to Combat Antimicrobial Resistance.
Drug Resistance in HIV
As noted earlier HIV is a retrovirus consisting of a single strand of RNA inside a protein coat. When HIV enters a CD4 lymphocyte, it sheds its protein coat and uses a viral enzyme called reverse trancriptase to create a segment of DNA using the viral RNA as a template. This double-stranded DNA version o HIV then gets incorporated into the DNA of the infected host cell, and this process is called "reverse transcription." This has important consequences for the development of drug resistance by HIV because of several key characteristics of HIV:
- HIV replicates at a prodigious rate producing billions of new virus particles each day.
- Reverse transcription is notoriously error prone, leading to frequent mutations.
- Current anti-viral treatment regimens control the infection, but they do not completely eliminate the virus.
Given the persistence of HIV with high rates of replication and high error rates during reverse transcription, mutations in HIV are inevitable, and some of these mutations lead to the eventual development of drug resistance. The video below summarizes the problem.
HIV infects CD4 lymphocytes and hijacks the cell's machinery to create viral proteins and RNA. HIV's genetic material in RNA, and in order replicate reverse transcriptase results in results in very rapid production of new HIV particles
Two concepts are important to an understanding of the development of drug resistance. First, HIV infection is characterized by high levels of virus production and turnover. In most untreated patients, the total number of productively infected cells in the lymphoid tissue has been estimated to be approximately 107 to 108cells. During the chronic phase of HIV infection, this number is relatively stable, reflecting the balance between the infection of new target cells and their clearance. Because the half-life of infected cells is remarkably short (one to two days), the maintenance of this steady state requires that HIV infect new target cells at a very high rate. Second, the viral population in an infected person is highly heterogeneous. The reverse transcription of viral RNA into DNA is notoriously prone to error,introducing on average one mutation for each viral genome transcribed. Most of these errors are base substitutions, but duplications, insertions, and recombination can also occur. The high rate of HIV infection, combined with the high mutation rate that occurs during each cycle of infection, ensures that patients have a complex and diverse mixture of viral quasispecies, each differing by one or more mutations.
Under these circumstances, it is easy to understand why if any of these mutations can confer some selective advantage to the virus, such as a decrease in its susceptibility to an antiretroviral agent, the corresponding quasispecies will overtake the others, following a simple darwinian selection process.