
Logistic Regression and Survival Analysis
Table of Contents»
Ching-Ti Liu, PhD, Associate Professor, Biostatistics
Jacqueline Milton, PhD, Clinical Assistant Professor, Biostatistics
Avery McIntosh, doctoral candidate
This module will enable you to perform logistic regression and survival analysis in R. The following topics will be introduced:
1. Why use logistic regression?
2. Overview of logistic regression.
3. How to perform logistic regression in R.
4. Why use survival analysis?
5. Overview of survival analysis (Kaplan-Meier plots and Cox regression)
6. How to perform survival analysis in R.
By the end of this session students will be able to:

Previously we discussed how to determine the association between two categorical variables (odds ratio, risk ratio, chi-square/Fisher test). Suppose we want to explore a situation in which the dependent variable is dichotomous (1/0, yes/no, case/control) and the independent variable is continuous.
Let's examine the Outbreak dataset in the epicalc library in R. Suppose we want to examine the association between vomiting (yes/no) as a dependent variable and age. Suppose we try to examine this association using linear regression.
E[ Vomiting ] = β0 + β1*age
If we run the linear regression model in R, what happens?
> library(epicalc)
> data(package="epicalc",Outbreak)
> attach(Outbreak)
> model.1<-lm(vomiting~age)
> summary(model.1)
> plot(model.1,which=1:4)

We see that just about all the assumptions of linear regression are violated in these plots. This is due to the fact that linear regression can only work well if the dependent variable is continuous. In this case the predicted values still fall between 0 and 1, which is good, but in many cases we can get an expected value of the probability of being a case that is above 1, or below 0; this is clearly erroneous.
When the assumptions of linear regression are violated, oftentimes researchers will transform the independent or dependent variables. In logistic regression the dependent variable is transformed using what is called the logit transformation:
![]()
Then the new logistic regression model becomes:
![]()
Covariates can be of any type:
and the outcome is binary: 0/1. Since Y is either 0 or 1, expected value of Y for a set of covariates X is thought of as "the probability that event Y occurs, given the covariates X." So we have that if p is the probability of vomiting, then
![]()
In linear regression, we were able to predict the outcome Y given new data by plugging in covariates on new data into the model. Here, we can do that for odd, odds ratios, or predicted probabilities (more on this later).
In standard linear regression, the coefficients are estimated based on the "least-squares" criterion. Here, the estimates for the coefficients are performed via Maximum Likelihood Estimation.
Due to a transformed outcome, there is a concomitant change in the interpretation of the regression coefficients:
log(odds of vomiting for those aged 20) = β0 + β1*20
log(odds of vomiting for those aged 21) = β0 + β1*21
odds of vomiting for those aged 20 = e β0 + β1*20
odds of vomiting for those aged 21 = e β0 + β1*21
If we want to determine the odds ratio to compare the odds of vomiting for those who are 20 years old versus the odds of vomiting for those who are 21 years old we can do the following:
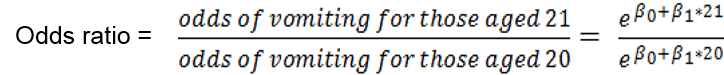
By laws of exponents, we can combine terms, and we get:
![]()
Thus ![]() = exp(β1) represents the odds ratio of vomiting comparing groups that differ by a one-unit change in age.
= exp(β1) represents the odds ratio of vomiting comparing groups that differ by a one-unit change in age.
If we want to get a predicted probability of the event of interest (p), given our covariates, then we can do a little algebra, and we have that
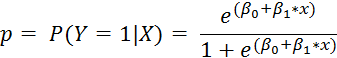
|
Generalized Linear Models |
|---|
|
There is an entire sub-field of statistical modeling called generalized linear models, where the outcome variable undergoes some transformation to enable the model to take the form of a linear combination, i.e. f (E[Y]) = β0 + β1X1+…+ βkXk. Logistic regression is just one such type of model; in this case, the function f (・) is f (E[Y]) = log[ y/(1 - y) ].
There is Poisson regression (count data), Gamma regression (outcome strictly greater than 0), Multinomial regression (multiple categorical outcomes), and many, many more. If you are interested in these topics, SPH offers · BS 853: Generalized Linear Models (logistic regression is just one class) · BS 820: Logistic Regression and Survival Analysis · BS 852: Statistical Methods in Epidemiology (covers some logistic and survival)
And in the math department there is · MA 575: Linear Models · MA 576: Generalized Linear Models |
To perform logistic regression in R, you need to use the glm() function. Here, glm stands for "general linear model." Suppose we want to run the above logistic regression model in R, we use the following command:
> summary( glm( vomiting ~ age, family = binomial(link = logit) ) )
Call:
glm(formula = vomiting ~ age, family = binomial(link = logit))
Deviance Residuals:
Min 1Q Median 3Q Max
-1.0671 -1.0174 -0.9365 1.3395 1.9196
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.141729 0.106206 -1.334 0.182
age -0.015437 0.003965 -3.893 9.89e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 1452.3 on 1093 degrees of freedom
Residual deviance: 1433.9 on 1092 degrees of freedom
AIC: 1437.9
Number of Fisher Scoring iterations: 4
To get the significance for the overall model we use the following command:
> 1-pchisq(1452.3-1433.9, 1093-1092)
[1] 1.79058e-05
The input to this test is:
This is analogous to the global F test for the overall significance of the model that comes automatically when we run the lm() command. This is testing the null hypothesis that the model is no better (in terms of likelihood) than a model fit with only the intercept term, i.e. that all beta terms are 0.
Thus the logistic model for these data is:
E[ odds(vomiting) ] = -0.14 – 0.02*age
This means that for a one-unit increase in age there is a 0.02 decrease in the log odds of vomiting. This can be translated to e-0.02 = 0.98. Groups of people in an age group one unit higher than a reference group have, on average, 0.98 times the odds of vomiting.
How do we test the association between vomiting and age?
When testing the null hypothesis that there is no association between vomiting and age we reject the null hypothesis at the 0.05 alpha level (z = -3.89, p-value = 9.89e-05).
On average, the odds of vomiting is 0.98 times that of identical subjects in an age group one unit smaller.
Finally, when we are looking at whether we should include a particular variable in our model (maybe it's a confounder), we can include it based on the "10% rule," where if the change in our estimate of interest changes more than 10% when we include the new covariate in the model, then we that new covariate in our model. When we do this in logistic regression, we compare the exponential of the betas, not the untransformed betas themselves!
|
Test the hypothesis that being nauseated was not associated with sex and age (hint: use a multiple logistic regression model). Test the overall hypothesis that there is no association between nausea and sex and age. Then test the individual main effects hypothesis (i.e. no association between sex and nausea after adjusting for age, and vice versa). |
Let's examine the dataset luekemia in the "survival" package.
> rm(list = ls())
> library(survival)
> data(leukemia)
> detach(Outbreak)
> data(package="survival",leukemia)
> attach(leukemia)
Survival in patients with Acute Myelogenous Leukemia. The question at the time was whether the standard course of chemotherapy should be extended ('maintenance') for additional cycles.
|
Variable |
Description |
|---|---|
|
time |
Survival or censoring time |
|
status |
censoring status (0 if an individual was censored, 1 otherwise) |
|
x |
was maintenance chemotherapy given? ("Maintained" if yes, "Not maintained" if no |
Suppose we want to examine the association between the length of survival of a patient (how long they survived leukemia) and whether or not chemotherapy was maintained. Suppose we create a histogram of the survival times.
> hist(time, xlab="Length of Survival Time", main="Histogram of Survial Time in AML Patients")
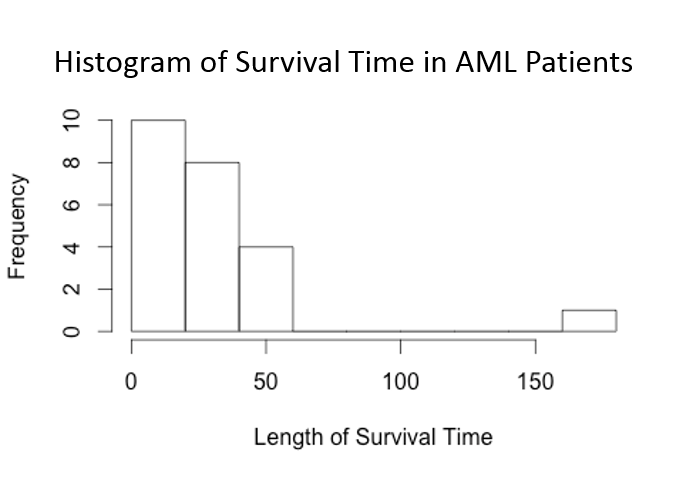
We see that the survival times are highly skewed due to the fact that there is a person who survived much longer than everyone else. In addition, there were quite a few people who survived for fewer than 10 years. This type of skewed distribution is typical when dealing with survival data and thus the normality assumption of linear regression is often violated, making it inappropriate to use.
In addition to this problem, we also see a problem known as censoring with survival data.
> leukemia[1:5,]
time status x
1 9 1 Maintained
2 13 1 Maintained
3 13 0 Maintained
4 18 1 Maintained
5 23 1 Maintained
The third observation has a status of 0. This means that the person was followed for 13 months and after that was lost to follow up. So we only know that the patient survived AT LEAST 13 months, but we have no other information available about the patient's status. This type of censoring (also known as "right censoring") makes linear regression an inappropriate way to analyze the data due to censoring bias.
One way to examine whether or not there is an association between chemotherapy maintenance and length of survival is to compare the survival distributions. This is done by comparing Kaplan-Meier plots.
> survfit(Surv(time, status)~1)
Call: survfit(formula = Surv(time, status) ~ 1)
records n.max n.start events median 0.95LCL 0.95UCL
23 23 23 18 27 18 45
This tells us that for the 23 people in the leukemia dataset, 18 people were uncensored (followed for the entire time, until occurrence of event) and among these 18 people there was a median survival time of 27 months (the median is used because of the skewed distribution of the data). The 95% confidence interval for the median survival time for the 18 uncensored individuals is (18, 45).
>summary(survfit(Surv(time, status)~1))
Call: survfit(formula = Surv(time, status) ~ 1)
time n.risk n.event survival std.err lower 95% CI upper 95% CI
5 23 2 0.9130 0.0588 0.8049 1.000
8 21 2 0.8261 0.0790 0.6848 0.996
9 19 1 0.7826 0.0860 0.6310 0.971
12 18 1 0.7391 0.0916 0.5798 0.942
13 17 1 0.6957 0.0959 0.5309 0.912
18 14 1 0.6460 0.1011 0.4753 0.878
23 13 2 0.5466 0.1073 0.3721 0.803
27 11 1 0.4969 0.1084 0.3240 0.762
30 9 1 0.4417 0.1095 0.2717 0.718
31 8 1 0.3865 0.1089 0.2225 0.671
33 7 1 0.3313 0.1064 0.1765 0.622
34 6 1 0.2761 0.1020 0.1338 0.569
43 5 1 0.2208 0.0954 0.0947 0.515
45 4 1 0.1656 0.0860 0.0598 0.458
48 2 1 0.0828 0.0727 0.0148 0.462
The following summary goes through each time point in the study in which an individual was lost to follow up or died and re-computes the total number of people still at risk (n.risk), the number of events at that time point (n.event), the proportion of individuals who survived up until that point (survival) and the standard error (std.err) and 95% confidence interval (lower 95% CI, upper 95% CI) for the proportion of individuals who survived at that point.
You can also plot the survival curves using the following commands:
> surv.aml <- survfit(Surv(time,status)~1)
> plot(surv.aml, main = "Plot of Survival Curve for AML Patients",
xlab = "Length of Survival", ylab = "Proportion of Individuals who have Survived")
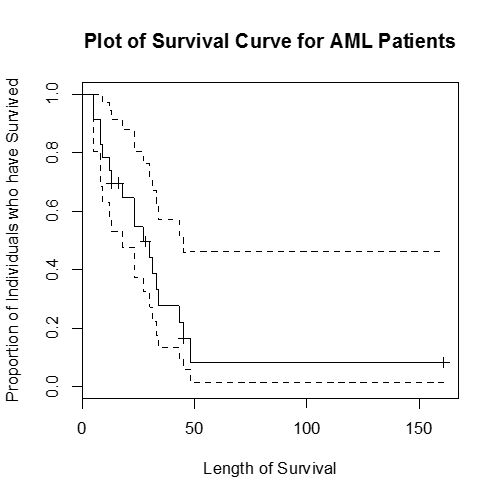
This plot shows the survival curve (also known as a Kaplan-Meier plot), the proportion of individual who have survived up until that particular time as a solid black line and the 95% confidence interval (the dashed lines).
Our original question was to examine the association between chemotherapy maintenance and length of survival. To do this in the context of survival analysis we compare the survival curves of those who received chemotherapy maintenance and those who did not. First, let's examine how to compare the survival statistics and create Kaplan-Meier plots for each chemotherapy group.
> print(survfit(Surv(time,status)~x))
Call: survfit(formula = Surv(time, status) ~ x)
records n.max n.start events median 0.95LCL 0.95UCL
x=Maintained 11 11 11 7 31 18 NA
x=Nonmaintained 12 12 12 11 23 8 NA
In this output we see that there is a total of 11 people who were on maintained chemotherapy, 7 died, the median follow up was 31. The 95% confidence interval of survival time for those on maintained chemotherapy is (18, NA); NA in this case means infinity. A 95% upper confidence limit of NA/infinity is common in survival analysis due to the fact that the data is skewed.
>plot(survfit(Surv(time,status)~x), main = "Plot of Survival Curves by Chemotherapy Group", xlab = "Length of Survival",ylab="Proportion of Individuals who have Survived",col=c("blue","red"))
>legend("topright", legend=c("Maintained", "Nonmaintained"),fill=c("blue","red"),bty="n")
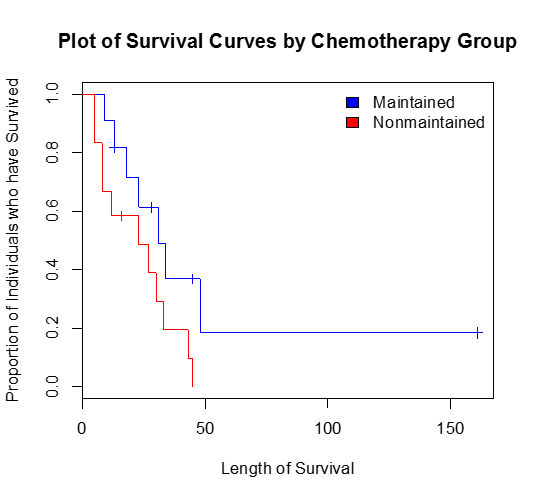
In order to determine if there is a statistically significant difference between the survival curves, we perform what is known as a log-rank test, which tests the following hypothesis:
The test is performed in R as follows:
> survdiff(Surv(time,status)~x)
Call:
survdiff(formula = Surv(time, status) ~ x)
N Observed Expected (O-E)^2/E (O-E)^2/V
x=Maintained 11 7 10.69 1.27 3.4
x=Nonmaintained 12 11 7.31 1.86 3.4
Chisq= 3.4 on 1 degrees of freedom, p= 0.0653
We see that when testing the null hypothesis that there is no difference in the survival function for those who were on chemotherapy maintenance versus those who were not on chemotherapy maintenance we fail to reject the null hypothesis χ12 = 3.4 with a p-value = 0.0653.
Suppose we want to see if there is a difference in survival functions between two groups after adjusting for a potential confounder.
We will not go into much detail on this model here, but briefly, a proportional hazard model is given as follows:
We define the hazard of an event as the risk of that event, as the time frame shrinks to 0. Usually when we calculate risk ratios, we have some time in mind, either cross-sectional, or, say, risk of dying after a year for two groups. Hazard is the risk, taken as the time frame vanishes to time t = 0.
![]()
h0(t) is the "baseline hazard," which we don't worry too much about, because when we look at the ratio of hazards for two conditions, we get the following:
Hazard ratio for individual with X = x vs. X = (x+1):
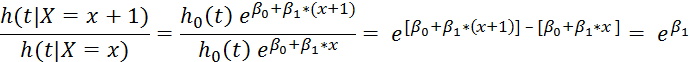
This term is the hazard ratio for the event of interest for people with covariate x+1 vs. people with covariate x. If the term is >1, then those people who have a one-unit increases in their covariate compared against a reference group are at a higher "risk" (hazard) for the event.
This is just the bare-bones basics of Cox Proportional Hazards models. There is a lot more to these models, including various assumptions that need to be tested for the model validity to hold, and issues in interpretation. However, for our purposes, just seeing how to run these models is enough.
Using a new dataset in the survival package called "cancer" we want to examine the survival in 228 patients with advanced lung cancer from the North Central Cancer Treatment Group. Performance scores rate how well the patient can perform usual daily activities.
|
Variable Name |
Description |
|---|---|
|
inst |
Institution code |
|
time |
Survival time in days |
|
status |
censoring status, 1=censored, 2=dead |
|
age |
age in years |
|
sex |
1=Male, 2= Female |
|
ph.ecog |
ECOG performance score (0= good, 5=dead) |
|
ph.karno |
Karnofsky performance score (0=bad, 100=good) rated by physician |
|
pat.karno |
Karnofsky performance score as rated by patient |
|
meal.cal |
calories consumed at meals |
|
wt.loss |
Weight loss in last six months |
First, let's do some non-adjusted analysis:
|
Test the null hypothesis that there is no difference in the survival function of patients with advanced lung cancer between males and females. |
Suppose we want to determine if there is an association between length of survival and gender after adjusting for age. The log-rank test discussed previously will only compare groups, it does not take into account adjusting for other covariates/confounding variables. To adjust for other covariates we perform the Cox's proportional hazards regression using the coxph() function in R. To perform exercise 2 using Cox regression we use the following commands:
> detach(leukemia)
> attach(cancer)
> coxph(Surv(time,status)~sex)
Call:
coxph(formula = Surv(time, status) ~ sex)
coef exp(coef) se(coef) z p
sex -0.531 0.588 0.167 -3.18 0.0015
Likelihood ratio test=10.6 on 1 df, p=0.00111 n= 228, number of events= 165
We see that when using the Cox regression to perform the test, the results are very similar to the log rank test (χ12 = 10.6 with p-value = 0.00111). The Cox regression estimates the hazard ratio of dying when comparing males to females. Here, sex is significantly related to survival (p-value = 0.00111), with better survival in females in comparison to males (hazard ratio of dying = 0.588). Females have 0.588 times the hazard of dying in comparison to males.
To extend the cox regression to adjust for other covariates, we will extend this to test the following hypothesis:
> coxph(formula = Surv(time, status) ~ sex + age)
Call:
coxph(formula = Surv(time, status) ~ sex + age)
coef exp(coef) se(coef) z p
sex -0.513 0.599 0.16746 -3.06 0.0022
age 0.017 1.017 0.00922 1.85 0.0650
Likelihood ratio test = 14.1 on 2 df, p=0.000857 n= 228, number of events= 165
When testing the null hypothesis that there is no difference in the survival function between males and females after adjusting for age we see that we reject the null hypothesis (z = -3.06, p-value = 0.0022). After adjusting for age, females have significantly better survival in comparison to males. Females have 0.599 times the hazard of dying in comparison to males, adjusting for age (HR<1).
Assignment:
|
For those who want to continue with R, SPH offers BS 845: Applied Statistical Modeling and Programming in R, taught by Professor Yang. |